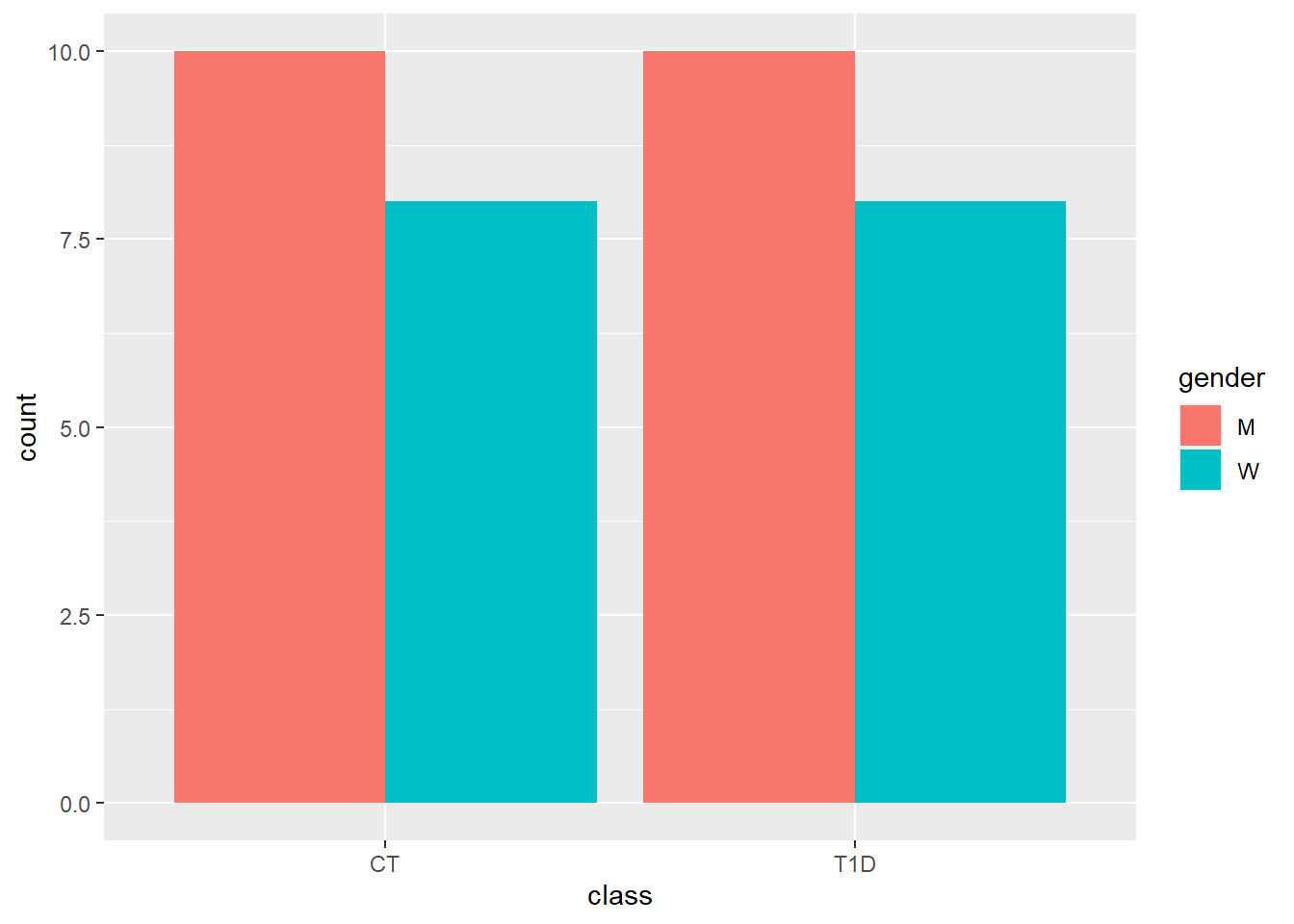
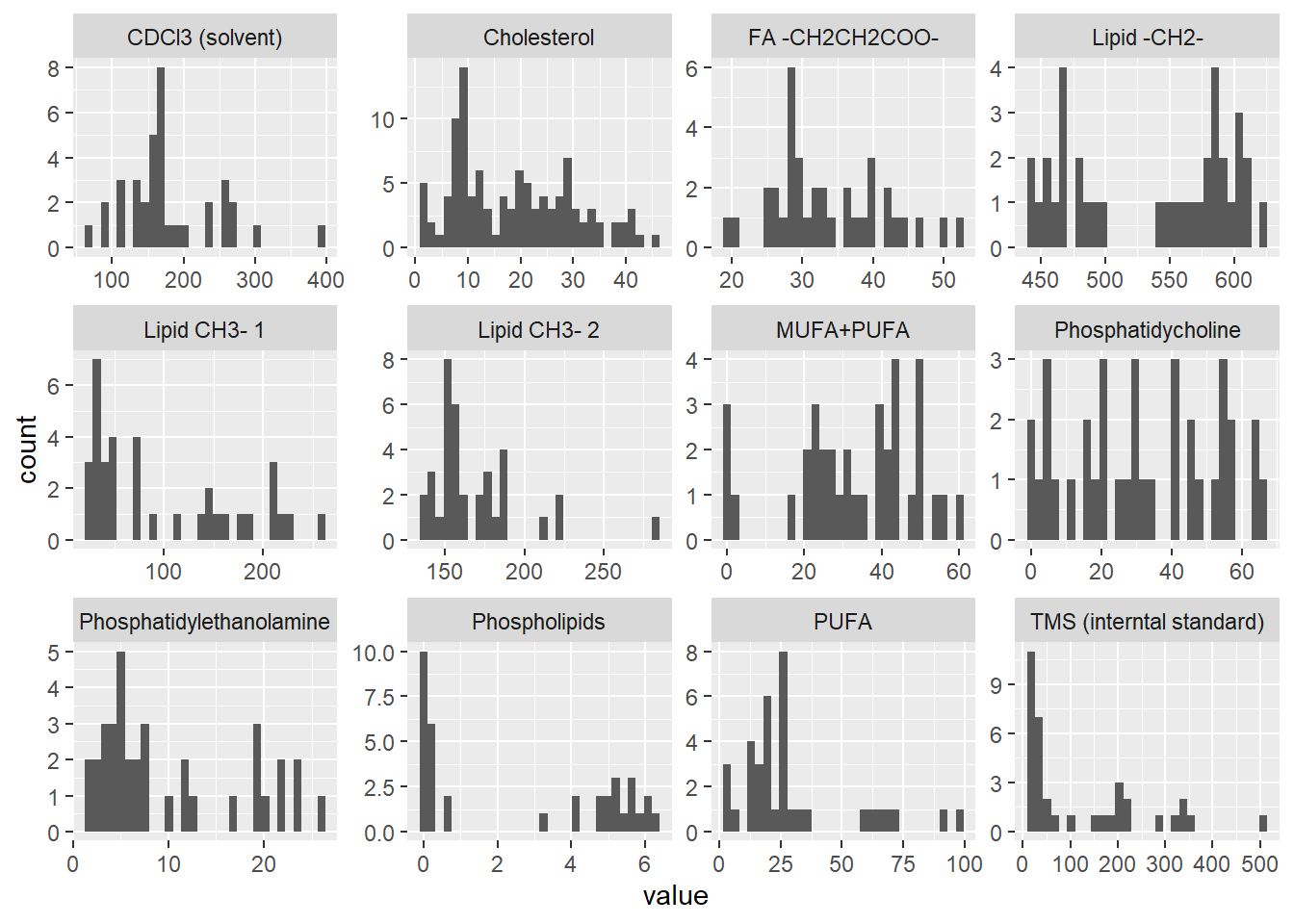
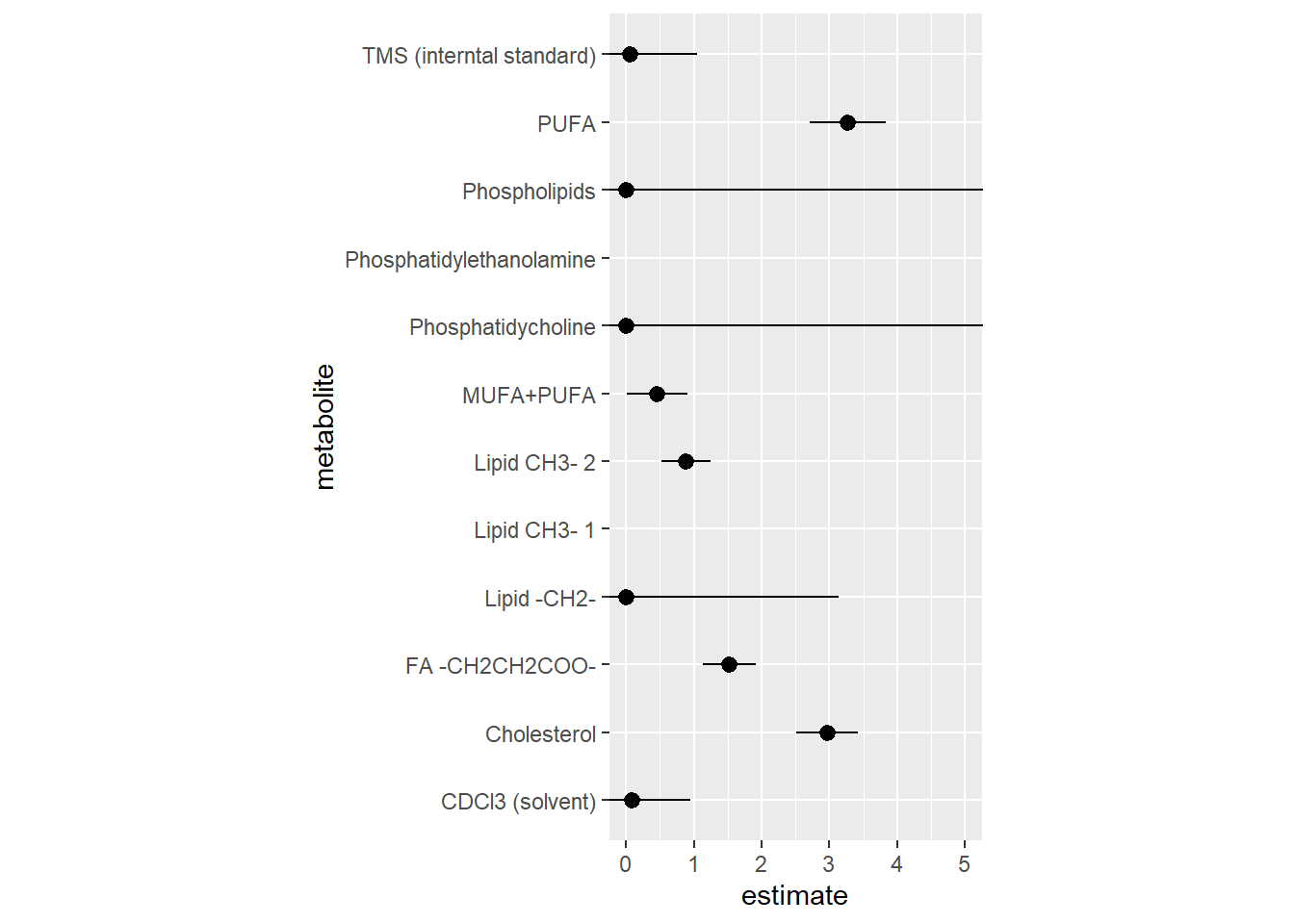
| metabolite | value_mean | value_sd | value_median | value_iqr | MeanSD |
|---|---|---|---|---|---|
| CDCl3 (solvent) | 180.0 | 67.0 | 166.7 | 70.9 | 180 (67) |
| Cholesterol | 18.6 | 11.4 | 17.1 | 19.0 | 18.6 (11.4) |
| FA -CH2CH2COO- | 33.6 | 7.8 | 31.4 | 11.3 | 33.6 (7.8) |
| Lipid -CH2- | 536.6 | 61.9 | 555.6 | 119.1 | 536.6 (61.9) |
| Lipid CH3- 1 | 98.3 | 73.8 | 72.7 | 122.4 | 98.3 (73.8) |
| Lipid CH3- 2 | 168.2 | 29.2 | 156.3 | 27.3 | 168.2 (29.2) |
| MUFA+PUFA | 32.9 | 16.1 | 34.5 | 21.3 | 32.9 (16.1) |
| Phosphatidycholine | 31.7 | 20.5 | 30.3 | 32.7 | 31.7 (20.5) |
| Phosphatidylethanolamine | 10.0 | 7.6 | 6.5 | 13.0 | 10 (7.6) |
| Phospholipids | 2.7 | 2.6 | 2.0 | 5.0 | 2.7 (2.6) |
| PUFA | 30.0 | 24.1 | 23.0 | 14.3 | 30 (24.1) |
| TMS (interntal standard) | 123.0 | 130.4 | 42.1 | 184.8 | 123 (130.4) |
Lipidomics analysis
Abstract
Objective:
Research Design and Methods:
Results:
Conclusions:
Introduction
Research Design and Methods
Results
# A tibble: 12 × 5
metabolite value_mean value_sd value_median value_iqr
<chr> <dbl> <dbl> <dbl> <dbl>
1 CDCl3 (solvent) 180 67 167. 70.9
2 Cholesterol 18.6 11.4 17.1 19
3 FA -CH2CH2COO- 33.6 7.8 31.4 11.3
4 Lipid -CH2- 537. 61.9 556. 119.
5 Lipid CH3- 1 98.3 73.8 72.7 122.
6 Lipid CH3- 2 168. 29.2 156. 27.3
7 MUFA+PUFA 32.9 16.1 34.5 21.3
8 Phosphatidycholine 31.7 20.5 30.3 32.7
9 Phosphatidylethanolamine 10 7.6 6.5 13
10 Phospholipids 2.7 2.6 2 5
11 PUFA 30 24.1 23 14.3
12 TMS (interntal standard) 123 130. 42.1 185.